ISC Biotechnology Previous Year Question Paper 2018 Solved for Class 12
Maximum Marks: 80
Time allowed: Three hours
- Candidates are allowed additional 15 minutes for only reading the paper. They must NOT start writing during this time.
- Answer Question 1 (Compulsory) from Part I and five questions from Part II, choosing two questions from Section A, two questions from Section B and one question from either Section A or Section B.
- The intended marks for questions or parts of questions are given in brackets [ ].
- Transactions should be recorded in the answer book.
- All calculations should be shown clearly.
- All working, including rough work, should be done on the same page as, and adjacent to the rest of the answer.
Part-I (20 Marks)
(Answer all questions)
Question 1.
(a) Mention any one significant difference between each of the following : [5]
(i) Plasmids and cosmids
(ii) Nucleotide and Nucleoside
(iii) Lagging strand and leading strand
(iv) Multipotent cells and unipotent cells
(v) Microinjection and biolistic
(b) Answer the following questions : [5]
(i) Who coined the term vitamin ? Write the chemical name of vitamin D.
(ii) Why is amino acid said to be amphoteric ?
(iii) What is Bioremediation ?
(iv) What is a primer ?
(v) What are cryoprotectants ?
(c) Write the full form of each of the following : [5]
(i) NBPGR
(ii) ARS
(iii) RELP
(iv) HEPA
(v) SCP
(d) Explain briefly : [5]
(i) Gene splicing
(ii) Supramolecular assembly
(iii) Interferon
(iv) Gene scan
(v) Saponification
Answer:
(a) (i) Plasmids and Cosmids
Plasmids: These are the extra-chromosomal, independent, self replicating, circular double stranded DNA molecules naturally found in all bacteria and some fungi.
Cosmids : These are predominantly plasmids with a bacterialori, an antibiotic selection marker and a cloning site, but they carry one or more recently two ‘cos’ sites derived from bacteriophage lambda.
(ii) Nucleotide and Nucleoside:
Nucleotide:
Nucleotide is a condensation product of heterocyclic nitrogen base, a pentose sugar like ribose or deoxyribose, and a phosphate or polyphosphate group.
Nucleoside:
Nucleoside consisting of a pentose sugar, usually ribose or deoxyribose, and a nitrogen base purine or pyrimidine.
(iii)Lagging strand and Leading strand
Lagging strand:
- Lagging strand is a replicated strand of DNA which is formed in short segments called Okazaki fragments. Its growth is discontinuous.
- DNA-ligase is required for joining Okazaki fragments.
Leading strand:
- It is a replicated strand of DNA which grows continuously without any gap.
- It does not require DNA ligase for its growth.
(iv) Multipotent cells and Unipotent cells
Multipotent Cells:
Multi potent cells have the ability to differentiate into many of the various types of specialized cell types and can develop into any cell of a particular group or type,
e.g., Umbilical cord stem cell
Unipotent Cells:
Uni potent cells can undergo unlimited reproductive divisions, but can only differentiate into a single type of cell or tissue.
e.g., Skin cells.
(v)Microinjection and biolistic
Microinjection:
In this technique foreign DNA is directly and forcibly injected into the nucleus of animal and plant cells through a glass micropipette containing very fine tip of about 0.5 mm diameter. It resembles with injection needle.
Biolistic:
In this technique macroscopic gold or tungsten particles are coated with desired DNA. The particles are bombarded onto target cells by the bombardment apparatus. Consequently foreign DNA is forcibly delivered into the host cells.
(b) (i) The term Vitamin was coined by Funk.
Chemical name of Vitamin D: Cholecalciferol (D3), Ergocalciferol (D2)
(ii) Amino acids are amphoteric, which means they have acidic and basic tendencies. The carboxyl group is able to lose a proton and the amine group is able to accept a proton.
Or
In neutral amino acid solution, the (– COOH) loses a proton and the – NH2 of the same molecule picks up the one. The resulting ion is dipolar charged but overall electrically neutral. Due to this reason amino acids are amphoteric in nature i.e., they donate or accept proton.
(iii) Bio-remediation : Nowadays, microbial preparations are used for degradation of these pollutants. The waste organic materials treated with selected potential strains of microorganisms are rendered into less or non-toxic forms. This process of remedying pollution is called bio-remediation.
(iv) Primer: A primer is a short strand of RNA that serves as a starting point for DNA synthesis. They are required for DNA replication because DNA polymerases can only add new nucleotide’s to an existing strand of DNA.
(v) Cryoprotectants: These are basically antifreeze that is added to the solutions in which the cells are being stored to protect them from membrane damage and ice crystal damage. Examples are DMSO, Polyethylene glycol.
(c) (i) NBPGR: National Bureau of Plant Genetic Resources.
(ii) ARS: Agricultural Research Service.
(iii) RFLP: Restriction Fragment Length Polymorphism
(iv) HEPA: High Efficiency Particulate Air Filter
(v) SCP: Single Cell Protein
(d) (i) Gene splicing: In molecular biology, splicing is a post transcriptional modification of an hnRNA, in which introns are removed and exons are joined.
(ii) Supra molecular assembly: It is based on weak bonds as supposed to covalent bonds. As mentioned weak bond, though harder to control have the advantage of possible reopening. Relying on weak bond in surface organisation, can lead to molecular assemblies and raise the probability of reorganization processes.
(iii) Interferon: They are natural anti-viral proteins produced by the cells of the system of most vertebrates in response to attack of foreign agents such as viruses, parasites and tumor cells. These belong to the large class of glycoprotein known as cytokine. Today interferon’s are used for the treatment of Hepatitis-C diseases.
(iv) Gene scan: It is a notable example of eukaryotic ab initio gene finders. Gene scan is one of the best gene finding algorithms for sequence alignment and gene prediction.
(v) Saponification : The hydrolysis of triacvlghcerol in the presence of sufficient sodium hydroxide is called saponification.
Part-II (50 Marks)
(Answer any five questions)
Question 2.
(a) Explain in detail, how Dolly, the sheep was created. [4]
(b) Mention any two chemical properties of each of the following: [4]
(i) Proteins
(ii) Carbohydrates
(c) What are Okazaki fragments ? How are they joined ? [2]
Answer:
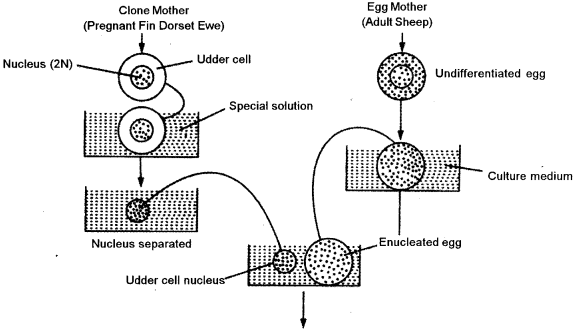
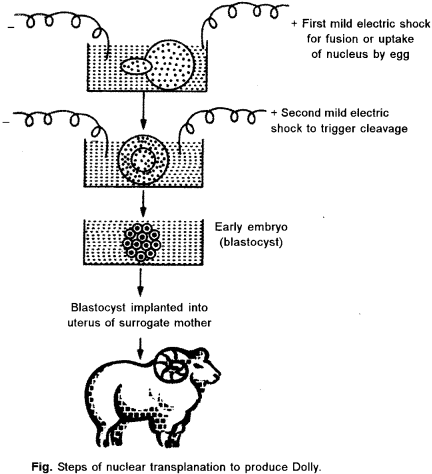
(a) Dolly, was the first mammal to be cloned by Wilmut et. al (1977). Dolly was the end result oi a long research program founded by the British government at the Roslin Institute in Scotland. They used the technique of somatic cell nuclear transfer, where the cell nucleus from an adult udder cell is transferred into an unfertilized oocyte that has had its nucleus removed. The hybrid cell is then stimulated to divide by an electric shock, and the blastocyst that is eventually produced’is implanted in a surrogate mother. Dolly was the first clone produced from a cell taken from an adult animal.
The following steps are involved in creation of Dolly:
Isolate donor nucleus : Isolate the nucleus from a somatic (non-reproductive) cell of a udder of adult donor sheep. The nucleus contains all the complete genetic material of the organism. A very small needle and syringe (suction device) is used to penetrate through the cell membrane to hold the nucleus and remove it from the cell.
Get unfertilized eggs: Retrieve some unfertilized egg cells (reproductive) from a female sheep. Many eggs are needed since not all of them will survive the various steps of cloning.
Remove the egg’s nucleus : Remove the egg cell’s nucleus, which contains only one-half of the sheep’s genetic material.
Insert donor nucleus : Insert the nucleus, with all its complete genetic material, isolated from the donor sheep mammal in Step 1 into the cytoplasm of egg cell that has no nuclear material. The egg’s genetic material now contains all traits from the donor adult. This egg is genetically identical to the donor adult cells.
Transfer the egg into womb : Transfer the egg into a receptive female sheep’s womb. Those eggs that survive and implant will continue to develop into embryos. When the off spring is born, it is a clone (genetically identical) of the donor sheep. After cloning was successfully demonstrated through the production of Dolly, many other large mammals have been cloned, including horses and bulls.
(b) Chemical properties of proteins:
- Proteins when hydrolyzed by acidic agents, like cone. HCl yield amino acids in the form of their hydrochlorides.
- Sanger’s reaction : Proteins react with FDNB reagent to produce yellow coloured derivative, DNB amino acid.
- Xanthoproteic test: On boiling proteins with cone. HNO3, yellow colour develops due to presence of benzene ring.
- Folin’s test: This is a specific test for tyrosine amino acid, where blue colour develops with phosphomolybdotungstic acid in alkaline solution due to presence of phenol group.
Chemical properties of carbohydrates:
Carbohydrates with free hemiacetals form (having anomeric carbon) are called reducing sugars such as glucose, fructose, maltose, etc., They can reduce alkaline solution of copper salts giving rise to yellow to red precipitate of cuprous oxides.
The reducing sugars give positive test for Fehling’s test, Benedict’s test and Barfoed s test. For the detection of sugars in blood/urine, these tests are carried out in a routine way in the pathology laboratory.
(c) Okazaki fragments are short, newly synthesized DNA fragments produced discontinuously in pieces during DNA replication. They are formed on the lagging template strand and are complementary to the lagging template strand. Okazaki fragments are joined together by DNA ligase enzyme.
Question 3.
(a) Describe the effect of each of the following factors on enzyme activity. [4]
(i) pH
(ii) Temperature
(iii) Enzyme concentration
(iv) Concentration of products
(b) With reference to suspension culture, explain the following : [4]
(i) A chemostat
(ii) A turbidostat
(c) What is genomics ? What are its different types ? [2]
Answer:
(a) (i) Optimum pH : Every enzyme has an optimum pH when it is most effective. A rise or fall in pH reduces enzyme activity 7 by changing the degree of ionization of its side chains. A change in pH may also start reverse reaction. Fumarase catalyses fumarate → malate at 6.2 pH and reverse at 7.5 pH. Most of the intracellular enzymes function near neutral pH with the exception of several digestive enzymes which work either in acidic range of pH or alkaline, e.g., 2.0 pH for pepsin, 8.5 for trypsin.
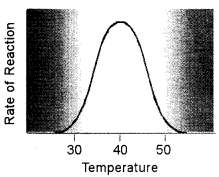
(ii) Temperature: Higher temperature generally causes more collisions among the molecules and therefore, increases the rate of a reaction. More collisions increase the likelihood that substrate will collide with the active site of the enzyme, thus increasing the rate of an enzyme-catalyzed reaction. Above a certain temperature, activity begins to decline because the enzyme begins to denature. The rate of chemical reactions, therefore, increases with temperature but then decreases as enzymes denature.
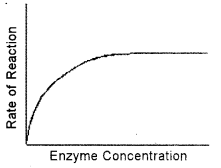
(iii) Enzyme Concentration: If there is insufficient enzyme present, the reaction will not proceed as fast as it otherwise would because all of the active sites are occupied with the reaction. Additional active sites could speed up the reaction. As the amount of enzyme is increased, the rate of reaction increases. This is so because when more enzyme molecules are present, more substrate molecules can be acted upon at the same time. This means that the total substrate molecules are broken down quickly. If there are more enzyme molecules than are needed, adding additional enzyme will not increase the rate. Reaction rate therefore increases as enzyme concentration increases but then its levels off.
(iv) Effect of Product Concentration: The accumulation of reaction products generally decreases the enzyme velocity. For certain enzymes, the products combine with the active site of enzyme and form a loose complex and thus, inhibit the enzyme activity. In the living system, this type of inhibition is generally prevented by a quick removal of products formed.
(b) (i) Chemostat : A type of cell culture; a component of medium is in a growth limiting concentration; fresh medium is added at regular intervals and equal volume of culture is withdrawn. But in a chemostat, a chosen nutrient is kept in a concentration so. that it is depleted very rapidly to become growth limiting, while other nutrients are still in concentrations higher than required. In such a situation, any addition of the growth- limiting nutrient is reflected in cell growth. Chemostats are ideal for the determination of effects of individual nutrients on cell growth and metabolism
(ii) Turbidostat : A type of suspension culture; when culture reaches a predetermined cell density, a volume of culture is replaced by fresh medium; works well at growth rates close to the maximum. A continuous culturing method where the turbidity of the culture is kept constant by manipulating the rate at which medium is fed. If the turbidity falls, the feed rate is lowered so that growth can restore the turbidity to its start point. If the turbidity rise • the feed rate is increased to dilute the turbidity back to its start point.
(c) Genomics: Genomics is an area within genetics that concerns the sequencing and analysis of an organism’s genome. The genome is the entire DNA content that is present within and cell of an organism. Experts in genomics strive to determine complete DNA sequences and perform genetic mapping to help understand disease.
Genome refers to the basic set of chromosomes. In a genome, each type of chromosome is represented only once. Now genomics is being developed as a sub discipline of genetics which is devoted to the mapping, sequencing and functional analysis of genomes.
Types of Genomics:
The discipline of genomics consists of two parts, viz. structural genomics and functional genomics.
Structural Genomics : It deals with the study of the structure of entire genome of a living organism. In other words, it deals with the study of the genetic structure of each chromosome of the genome. It determines the size of the genome of a species in mega-bases [Mb] and also the genes present in the entire genome of a species.
Functional Genomics: The study of function of all genes present in the entire in the genome is known as functional genomics. It deals with transcriptome and proteome. The transcriptome refers to complete set of RNA’s transcribed from a genome and proteome refers to complete set of proteins encoded by a genome.
Question 4.
(a) What are the basic facilities that should be available for tissue culture in a biotechnology laboratory ? [4]
(b) Explain the experiment which proves the semi-conservative mode of replication. [4]
(c) What is cDNA ? [2]
Answer:
(a) A biotechnology laboratory is a place for working with a variety of chemicals and desired organisms since several culture media are prepared and organic materials are present, their exists chance for the presence of high spectrum of microbial community.
Facilities and Tools for Tissue culture in a Biotechnology’ Laboratory:
A biotechnology laboratory should be well-equipped with the instruments. Plant tissue culture can be defined as a collection of techniques/methodologies that are employed to grow or maintain plant cells, tissues or organs on a nutrient culture medium under aseptic growth conditions. Micro propagation is a technique which is widely used for the mass production of clones of a plant. Due to its diverse applications in reviving/ maintaining plants this technique has immensely benefited our society. Therefore, it becomes important to understand the requirements to start tissue culture laboratory.
Some of the prerequisite basic facilities for any laboratory, which actively practices the plant tissue culture methods to grow plants, are :
- Washing area
- Glassware
- Autoclave
- Areas for media preparation, sterilization, and storage
- Area for transfer of cultures/medium in aseptic conditions.
- Environmentally controlled incubators or culture rooms.

Biotechnology equipment available for all major research laboratory’ techniques, including:
- Electrophoresis and blotting.
- Imaging and quantitation.
- Real-time PCR and digital PCR.
- Transfection.
- Flow cytometry, cell sorting, and cell counting.
- Preparative and analytical chromatography.
- Spectrophotometry and fluorometry.

(b) In 1958, for the first time M.S. Messelson and F. W. Stahl put experimental evidences for replication of DNA in a semi-conservative way as proposed by Waston and Crick (Fig). They grew the cells of Escherichia coli in the medium containing heavy’ isotopic nitrogen (15N) for about 14 cell generations. This was done with the objective that all light isotope nitrogen (14N) of purine and pyrimidine could be replaced by the isotopic heavier nitrogen i.e., 15N. The cells were taken out the DNA of cells was removed. The cells grown on 15N containing medium were filtered and properly w’ashed. Density of 15N – DNA w as heavier (1.772 g/cc) as compared to normal DNA (1.708 g/cc).
Again the cells grown on 15N containing medium were taken out, washed properly and grown on medium containing light isotopic nitrogen (14N) for one to two generations. The generation time of E.coli is 30 minutes. Therefore, it was possible to remove cells after 30 minutes or 60 minutes. Density of 14N-DNA of such cells was measured by following density gradient centrifugation. Explanation of the result obtained by Messelson and Stahl can be summarized as below:
After 14 generation all the 14N of bases were replaced by 15N. This is why the density of 15N DNAwas higher.
DNA sample analysed after one cell generation transferred to 14N – containing medium showed only one density band when observed under ultra violet light. The DNA contained 50% each of 15N and 14N and, therefore, the density 7 was intermediate between the density of heavy DNA and light DNA.
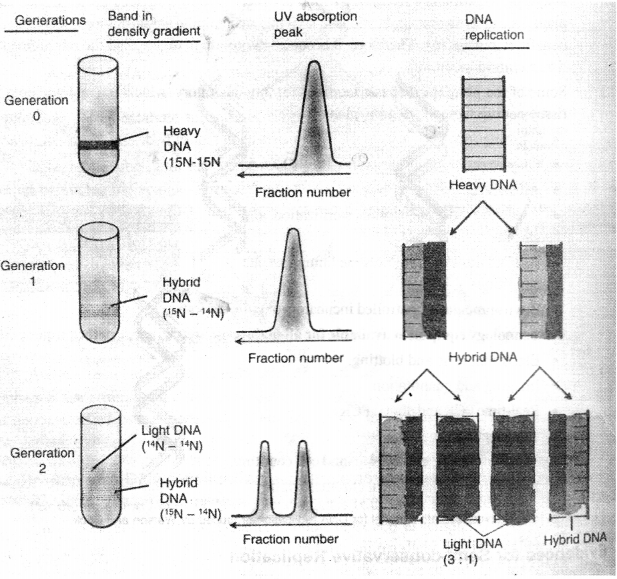
DNA analysed after two generations of E. coli cells showed the presence of two bands due to types of DNA helix. One type consisted of 15N – 15N hybrid and the second were made- up of only 15N/.e., 14N- 14N. The ratio of heavy and light DNAwas 3 : 1. Moreover, the ratio of heavy DNA is subsequent generations gradually decreased and that of light DNA increased. This experiment shows that DNA replicates by semi-conservative method. Thereafter, several evidences for semi-conservative way of DNA replication were given by many scientists such as J. Cairns (1962) and J.HTylor (1957) using auto radigraphic techniques is E. coli and Vicia fab a. respectively
(c) cDNA : The double stranded molecules prepared by using mRNA as template and reverse transcriptase are called cDNA. These are expressed genes of the genomic DNA. By using cDNA molecules, substantial number of sequences have been determined and deposited in database.
Question 5.
(a) Explain any four methods employed to induce haploid production. [4]
(b) Describe the automated method of DNA sequencing. [4]
(c) What is the difference between gel electrophoresis and gel permeation? [2]
Answer:
(a) In vitro culture of haploid cells of plants {e.g., pollen grains from anther and ovules from ovary) is possible. In vivo technique of haploid production includes the following :
- Androgenesis: Production of haploid plants by development of an egg cell containing male nucleus. The female nucleus is eliminated before fertilisation.
- Gymnogenesis: Production of haploid plant by the development of an unfertilised egg cells because of delayed pollination (through use of abortive pollen pre-exposed to ionising irradiation or using an alien pollen). It is found in interspecific crosses of potato.
- Chemical Treatment: Some chemicals like chloramphenicol and parafluorophenylalanin may induce chromosome elimination in somatic cells or tissues giving rise to haploid.
In 1953, Tulecke successfully produced callus from isolated pollen culture of gymnosperms. This made a hope to raise haploid plants through pollen culture. For the first time Hameya and Hinata (1970) produced tissues from the isolated pollen grains of an angiosperm.
For the first time S.Guha and P.Maheshwari (1964) produced haploid embryos in vitro from isolated anthers of Datura innoxia. Haploid production has immense use in plant breeding and improvement of crop plants. Haploids provide an easier system for induction of mutation. They can be employed for rapid selection of mutants having traits for disease resistance. The Institute of Crop Breeding and Cultivation (China) has developed the high yielding and blast resistant varieties of rice zhonghua No. 8 and zhonghua No. 9 through transfer of desired alien gene.
(b) Automatic DNA Sequencers: Automatic sequencing machines were developed during 1990’s. It is an improvement of Sanger’s method. In this new method, a different fluorescent dye is tagged to the aWNTPs. Using this technique, a DNA sequence containing thousands of nucleotide’s can be determined in a few horns. Each dideoxynucleotide is linked with a fluorescent dye that imparts different colours to all the fragments terminating in that nucleotide. All four labelled ddNTP’s are added to a single capillary tube. It is a refinement of gel electrophoresis which separates fastly. DNA fragments of different colours are separated by their size in a single electrophoretic gel.
A current is applied to the gel. The negatively charged DNA strands migrate through the pores of gel towards the positive end. The small sized DNA fragments migrate faster and vice-versa. All fragments of a given length migrate in a single peak. The DNA fragments are illuminated with a laser beam. Then the fluorescent dyes are excited and emit light of specific wavelengths which is recorded by a special ‘recorder’. The DNA sequences are read by determining the sequence of the colours emitted from specific peaks as they pass the detector. This information is fed directly to a computer which determines the sequence. A tracing electrogram of emitted light of the four dyes is generated by the computer. Colour of each dya represents the different nucleotides. Computer converts the data of emitted light in the nucleotide sequences.
(c) Gel electrophoresis is a technique by which negatively charged DNA fragments are separated by forcing them to move towards the anode under an electric field through a medium/matrix. Gel permeation or filtration involves that molecules of different sizes can be separated from each other on the basis of their ability to enter the pores within the beaded gel, followed by passing down a column containing the gel. The technique is used in protein purification.
Question 6.
(a) What is in vitro pollination ? Why is it done ? Write the steps involved in this process. [4]
(b) Why is Agrobacterium called a natural genetic engineer ? How does it help in creating transgenic plants ? [4]
(c) Write a short note on site directed mutagenesis. [2]
Answer:
(a) In vitro Pollination : Cultivation of plant tissue or other organs on artificial media in a test tube or conical flask is called in vitro technique. The process of seed formation following stigmatic pollination of cultured pistil has been referred to as in vitro pollination.
Need of Invitro Pollination:
- For the production of homozygous plant
- For the conservation of extinct plant species
- Hybrid production
- Reducing the breeding cycle
- Overcome the dormant period
- Production of haploid plant
- Conservation of germplasm
(b) The bacterium Agrobacterium tumefaciens contains inside it s cell, a plasmid, i.e., an extrachromosomal self-replicating small DNA, known as Ti plasmid (tumour including plasmid) that induce overproduction of growth factors in the host cells iventually leading to cell proliferation and tumour formation. This bacterium establishes contact with suitable root cells of plants by recognising chemical signals and then prepares it’s plasmid ready to be delivered through t-pilus developed by type IV secretion system. Then it transfers this DNA (also known as t-DNA) into the plant host cell. This delivery requires assistance of several virproteins.
The t-DNA is transferred and stably integrated into the host chromosome which later wards leads in tumour development in the host. The whole process of successful DNA transfer and integration is performed and monitored by the bacteria itself (which in vivo is done by genetic engineers in laboratories). That’s -why Agrobacterium tumefaciens is called the Natural genetic engineers of plants.
Role of Agrobacterium tumefaciens in the production of Transgenic Plants:
Genetically Modified Organisms (GMO’s) are organisms which are formed when the gene of interest is specifically inserted into the host organisms in order to alter its genome and produce an organism with the required desirable traits.
Agrobacterium tumefaciens is a bacterium which naturally causes tumors in plants which is caused by the transfer of DNA (Deoxyribonucleic Acid) from the bacterium to the plant it infects.
Therefore, in the production of transgenic plants by A. tumefaciens, plants do not acquire different traits as A. tumefaciens is used to transfer genes so that the plants acquire the traits. Also, neither resistance is given to plants by this bacterium nor plant genes are incorporated into the genome of this bacterium in the production of transgenic plants.
(c) Site – Directed Mutagenesis: In nature, mutations arise spontaneously but they are rare such as sickle cell anemia. The most powerful method of introducing point mutation into a gene is known as Oligonucleotide or site-directed mutagenesis.
Change in a single nucleotide base pair is called “point mutation”. Site-directed mutagenesis is a molecular biology technique in which a mutation is created at a specific site in the DNA molecule. Using this technique specific point of a gene can be mutated. Therefore, this method has been used to understand the function of many genes. Moreover, this technique can only be used when nucleotide sequence of gene is known.
Question 7.
(a) What is HGP ? Name any two scientists involved in this. Write any two achievements of HGP. [4] (h) List the functions of the following in Bioinformatics [4]
(i) ENTREZ
(ii) PDB
(iii) FASTA
(iv) MGD
(c) Mention two differences between the organisation of prokaryotic and eukaryotic genomes. [2]
Answer:
(a) (i) HGP: HGP means Human Genome Project. It was officially started on October. 1990 in the USA. This is an International Scientific Research Project with a primary goal to determine the sequence of chemical base pairs which make-up DNA and to identify approximately 25000 genes of the human genome from both a physical and functional standpoint.
Significance of genomics was substantially increased when the Human Genome Project was conceived. Research work on Human Genome Project was successfully completed only due to international collaboration involving 60 countries, 20 Genome Research Centers and more than 1000 scientists.
The US side of the Human Genome Project was initially led by James Watson (one half of Crick and Watson, who discovered the structure of DNA), and later by Francis Collins. John Sulston, who was the director here at the Welcome Trust Sanger Institute (at that time called the Sanger Center), was principal leader of the UK side of the project.
Achievements of HGP:
The main objective of this project was to find the full structure of human genes. After that the human genome project kept the target in front of it to know about the protein made by human genes. As a result it would be possible to find out the presence, decreasing number and faults of genes and likewise it would be possible to develop the process of treatment of the diseases like cancer, diabetes, AIDS etc. Along with this, the progress in the treatment technique is also and objective of this project.
The greatest achievement of this project is that scientists can find out the structure of human genes and successfully trace the structure of genes. It has been possible to develop the technique regarding the treatment of defective genes only because of the human genome project.
The treatment of the hereditary’ diseases has been easier because of this project. DNA interference is recently developed technique regarding treatment by which it would be possible to treat many uncureable diseases. Along with this, the development of human, phy sical and mental structure etc., are the main achievements of human genome project.
(b) (i) ENTREZ: Integrated information database retrieval system of NCBI. Using Entrez system we can access literature, sequences (both proteins and nucleotides) and structures (3D).
(ii) PDB: (Protein Data Bank): This database has sequence of those protein whose 3-D structures are known Sources: NCBI – USA; EBI, U.K.
(iii) FASTA:
- FASTA is a DNA and protein sequence alignment software package.
- The current FASTA package contains programs for protein: protein, DNA: DNA, protein: translated DNA (with frame shifts), and ordered or unordered peptide searches. Recent versions of the FASTA package include special translated search algorithms that correctly handle frame shift errors (which six-frame-translated searches do not handle very well) when comparing nucleotide to protein sequence data.
(iv) Mouse Genome Database:
MGD strives to provide an extensively integrated information resource with experimental details annotated from both literature and on-line genomic data sources.
MGD collaborates with other bioinformatics groups to curate a definitive set of information about the laboratory mouse.
(c)
| Prokaryotic Genome | Eukaryotic Genome |
| (i) Genome are much smaller and simpler. | (i) Genome are larger and complex. |
| (ii) Highly repetitive DNA is not found. | (ii) Occurrence of highly repetitive DNA is found. |
| (iii) It is without a limiting membrane. | (iii) It is bounded by nuclear membrane. |
| (iv) It is a naked double strand of DNA. | (iv) Double strand of DNA is associated with histone proteins |
Question 8.
(a) Briefly describe the steps involved in the Southern blotting technique. [4]
(b) What is the need of germplasm conservation ? Give an account of the in-situ and ex-situ conservation of germplasm. [4]
(c) What is peptidoglycan ? Where is it found ? [2]
Answer:
(a) Southern blotting is a technique for transfer of DNA molecules from an electrophoresis gel to a nitrocellulose or nylon membrane, and is carried out prior to detection of specific molecules by hybridization probing.
In this technique, DNA is usually converted into conveniently sized fragments by restriction digestion and separated by gel electrophoresis, usually on an agarose gel. The DNA is denatured into single strands by incubation by alkali treatment.
The DNA is transferred to a nitrocellulose filter membrane which is a sheet of special blotting paper. The DNA fragments retain the same pattern of separation they had on the gel. This process is called blotting.
The nitrocellulose membrane is how removed from the blotting stack.
The blot is incubated with many copies of a radioactive probe which is single-stranded DNA. This probe detect and identify base pairs with its complementary DNA sequence and bind to form a double-stranded DNA molecule. The probe cannot be seen but it is either radioactive or has an enzyme bound to it (e.g., alkaline phosphatase or horseradish peroxidase). This step is known as hybridisation reaction.
The location of the probe is revealed by incubating it with a colourless substrate that the attached enzyme converts to a coloured product that can be seen or gives off light which will expose X-ray film. If the probe was labelled with radioactivity, it can expose X-ray filmdirectly. The images of radioactive probe are revealed as distinct bands on the developed X-ray film.
(b) Germplasm Conservation: The sum total of all the genes present in a crop plant and its related species is called germplasm . The wild and traditional species must be preserved through germplasm conservation.
Importance or Need of Germplasm Conservation:
Due to rapidly growing human population, plant and agriculture scientists have produced many new varieties to feed the population. Because the traditional and wild varieties cannot meet the demand of increasing population. But the traditional varieties need to be preserved because they are important for future breeding programmes. However, there is danger of erosion of genetic resources due to extensive use of newly introduced varieties.
In 1972, conservation of habitats rich in genetic diversity was recommended in the UN conference. Then an International Board for Plant Genetic Resource (IBPGR) was established. This board has objectives to provide necessary support for collection, conservation and utilisation of plant genetic resources from anywhere in the world. Germplasm preservation using cell and tissue cultures is done with the following objectives :
- Conservation of somaclonal and gametoclonal variation in cultures,
- Maintenance of recalcitrant seeds,
- Conservation of cell lines producing medicines,
- Storage of pollen for enchancing longevity,
- Conservation of rate germplasm arising through somatic hybridisation.
- Delaying the process of ageing,
- Storage of meristem culture for micropropagation, micrografting and production of disease- free plants,
- Conservation of plant materials from endangered species,
- Establishment of germplasm bank, and
- Exchange of information and germplasm at international level.
Modes of Conservation : It has been estimated that about 9,000 wild plant species are threatened. Global climatic changes also affect the natural plant habitats, thereby contributing to rapid changes in agricultural strategies. There are two modes of conservation of plant species as given below:
(a) In situ Conservation : Since 1980, in situ conservation has received high priority in the world conservation strategy. The method of conservation is to preserve land races with wild relatives in which genetic diversity exists.
(b) Ex situ Conservation: It is the chief mode of conservation of genetic resources including both cultivated and wild ones. Under suitable conditions genetic resources are conserved for a long term as gene bank.
(c) Peptidoglycan : Peptidoglycan, also called murein, is a polymer that makes up the cell wall of most bacteria. It is made-up of sugars and amino acids, and when many molecules of peptidoglycan joined together, they form an orderly cry stal lattice structure. Peptidoglycan is the main component of the Cell wall in most bacteria. Cross-linking between amino acids in the layer of peptidoglycan forms a strong meshlike structure that provides structure to the cell. Peptidoglycan provides a very important role in bacteria because bacteria are unicellular; it gives strength to the outer structure of the organism.
Question 9.
(a) How are biomolecules separated by the following techniques : [4]
(i) Ion exchange chromatography
(ii) Partition chromatography.
(b) What is the cause and the symptoms of the following diseases : [4]
(i) Sickle cell anaemia
(ii) Alkaptonuiria
(c) What is the difference between peptide bond and phosphodiester bond ? [2]
Answer:
(a) (i) Ion Exchange Chromatography: It is based on reversible exchange of ions in solution with ions electrostatically bound to some sort of insoluble support medium. Separation is obtained since different molecules have different degree of interaction with ion exchanger due to difference in their charges, charge densities and distribution of charge on their surface.
(ii) Partition Chromatography: Partition chromatography is the process of separation whereby the components of the mixture get distributed into two liquid phases due to differences in partition coefficients during the flow of mobile phase in the chromatography column. Here, the molecules get preferential separation in between two phases, i.e., both stationary phase and mobile phase are liquid in nature. So molecules get dispersed into either phases preferentially. Polar molecules get partitioned into polar phase and vice-versa. This mode of partition chromatography applies to liquid-liquid, liquid-gas chromatography and not to solid-gas chromatography. Because partition is the phenomenon in between a liquid and liquid or liquid and gas or gas and gas. But not in solid involvement.
(b) (i) Sickle cell anaemia : It is an autosomal hereditary disorder due to a mutation of single nitrogen base. It results in the formation of an abnormal haemoglobin called haemoglobin S (Hbs). In this, only one amino acid-6th amino acid of β-chain glutamic acid is replaced by valine. The erythrocytes become sickle shaped under oxygen deficiency as during strenous exercise and at high altitudes. They cannot pass through narrow capillaries. They clog blood capillaries. Blood circulation and oxygen supply is disturbed. Spleen and brain get damaged. Patient feels acute weakness. Homozygotes (Hbs / Hbs) usually die before reaching maturity.
(ii) Alkaptonuria: This was one of the first metabolic diseases described by Garrod in 1908. It is an inherited metabolic disorder produced due to deficiency of an oxidase enzyme required for breakdown of homogentisic acid (acid called alcapton, hence, alkaptonuria is also written as alcaptonuria.) Lack of the enzyme is due to the absence of the normal form of gene that controls the synthesis of the enzyme. Hence, homogentisic acid then accumulates in the tissues and is also excreted in the urine. The most commonly affected tissues are cartilages, capsules of joints, ligaments and tendons. The urine of these patients if allowed to stand for some hours in air, turns black due to oxidation of homogentisic acid.
(iii) Peptide bond:
A peptide bond is a covalent bond formed between two amino acids. Living organisms use peptide bonds to form long chains of amino acids, known as proteins. Proteins are used in many roles including structural support, catalyzing important reactions, and recognizing molecules in the environment. A peptide bond is therefore the basis of most biological reactions.
Phosphodiester bond:
A phosphodiester bond occurs when exactly two of the hydroxyl groups in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. Phosphodiester bonds are central to all life on Earth, as they make up the backbone of the strands of nucleic acid.